Introduction
Colorectal cancer (CRC) is one of the most common malignant
tumor with high migration and invasion capacity. It is estimated
that accounting for 9.0% of deaths worldwide [1]. In 2020, the
number of new cases of colorectal cancer in China exceeded
550,000 and the number of deaths exceeded 280,000 [2]. Despite
advancements in treatment over the few past decades, the 5-year
survival rate of Metastatic CRC was still under 12%. Several factors
were reported to influence CRC prognosis, such as lifestyle [3],
genome instability [4] aberrant gene expression, etc. However,
the mechanism underlying CRC survival remains elusive, which
impedes the improvement of CRC prognosis.
ZNF692 also known as AREBP or Zfp692 is located on chromosome 1q44. This gene has 30 transcripts (splice variants), 136 orthologues and 36 paralogues and ZNF692 protein was first identi-
fied as a transcription factor bound to the promoter elements of
phosphoenolpyruvate carboxykinase [5]. Recent research showed
that ZNF692 has different RNA splicing events within various
types of carcinoma [6,7]. Additionally, overexpression of ZNF692
has been reported to be related with the worse overall survival of
lung adenocarcinoma [8]. However, the expression profiles and
molecular functions of ZNF692 in CRC remain unclear.
The Cancer Genome Atlas (TCGA) is a public funded project
that aims to catalog and discover major cancer-causing genomic
alterations [9,10]. So far, TCGA researchers have analyzed large
cohorts of over 30 human tumors through large scale genome
sequencing and integrated multi-dimensional analyzes. Studies
of individual cancer types have extended current knowledge of
tumorigenesis.
In this study, a total of 248 CRC patients were enrolled. Table
1 presents the relationship between clinical factors and ZNF692
expression in patients with CRC. Age, M stage, and histological
grade were significantly associated with ZNF692 expression. The
proportion of high expression of ZNF692 for younger patients
was significantly greater than older patients (62.7% vs. 42.0%,
P=0.001). M1 stage patients with a higher rate for high expression
of ZNF692 compared with M0 stage patients (68.4% vs. 48.1%,
P=0.021). Middle/high grade patients showed a higher rate of
ZNF692 high expression (55.8% vs. 38.6%, P=0.021). Relative
higher expression of ZNF692 was associated with inferior overall
survival. Furthermore, ZNF692 expression was proved to be an
independent negative prognosis factor for Overall Survival (OS) in
multivariate survival analysis as shown in table 2. Moreover, the
adverse effect of high expression of ZNF692 on the prognosis of
patients was also verified by TCGA data analysis of CRC.
Materials and methods
Patients
The colorectal cancer specimens involved in this study are all
selected from patients undergoing surgical treatment in the Affiliated Hospital of Jiangnan University in China from August 2013
to December 2014. Case inclusion criteria: The primary tumor
specimen was colorectal cancer, and all specimens were verified
by two pathologists after the operation. The patients did not receive radiotherapy or chemotherapy before surgery. There are
complete clinical, pathological and follow-up data, the cause of
death is only tumor recurrence or metastasis. There was no second malignant tumor other than colorectal cancer within 5 years.
Here, a total of 248 patients met the above criteria. There were
133 males and 115 females; with an average age of 63 years; According to the 8th edition of the AJCC colorectal cancer staging,
there are a total of 29 cases in stage I , 99 cases in stage II, 82
cases in stage III, and 38 cases in stage IV. This study was approved
by the medical ethics committee of our hospital, and all patients
gave informed consent.
Tissue microarray (TMA) and immunohistochemistry staining
(IHC)
The method of TMA and IHC were carried out in accordance
with our previous standard operations [11]. ZNF692 polyclonal antibodies were purchased from Beijing Boaosen Technology
(Cat: bs-4360R), GTVisionTM III anti-rat/rabbit universal immunohistochemistry detection kit were purchased from Shanghai Gene
Technology (GK500705), tissue chip blank wax blocks were purchased from UNITMA Quick-Ray (UB06-1, South Korea). Perforate
the corresponding part marked on the donor wax block to collect
the tissue core, with a diameter of 2 mm, transfer the tissue core
to the hole of the acceptor module, and perforate each specimen
twice. Then perform routine immunohistochemistry sectioning,
staining and scoring. Briefly, sections of formalin-fixed, paraffin-embedded tissue were immunostained using ZNF692 polyclonal
antibodies. The staining results revealed that ZNF692 was positively located in the nucleus and cytoplasm. Score by staining
intensity and percentage of stained cells. A score of 0(no staining), 1(the tumor cells are light yellow stained without obvious
granules or less than 10% of the tumor cells are yellow stained
with obvious granules), 2(more than 10% of tumor cells are yellow stained with obvious granules or less than 10% of tumor cells
are brown stained with obvious granules), 3(More than 10% of
tumor cells are brown stained with obvious granules) within carcinomatous areas. A score of <2 is judged as low expression, and a
score ≥2 is judged as high expression. Results were independently
double-blind reading assessment by physicians in the pathology
department.
Bioinformatics analysis
The data of 512 colorectal cancer patients (COAD) in TCGA
(The Cancer Genome Atlas) were downloaded using UCSC Xena
(https://xenabrowser.net/datapages/), by which the genes differential expression analysis was also conducted. The genes with FDR
0.05 and |log FC| 1 were defined as differentially expressed genes
(DEGs) and were further analyzed (R software 4.1.2). Overall survival of TCGA-COAD patients was determined by Kaplan-Meier
analysis in 487 patients with survival data. Hazard ratio (HR) and
corresponding 95% confidence interval (CI) was calculated with
an optimal cutoff value, and log-rank P<0.05 was considered statistically significant for a difference.
Statistical analysis
Statistical analyses were conducted using SPSS, version 18.0.
We performed Chi-square tests to examine the associations between ZNF692 expression and clinical characteristics. Hazard ratios (HRs) and corresponding 95% confidence intervals (CIs) for
association of clinical characteristics and overall survival (OS)
were estimated using univariate and multivariate survival analysis
using Cox’s regression model respectively. Differences were considered statistically significant at P <0.05.
Results
Expression of ZNF692 in colorectal cancer tissues
By immunohistochemistry staining, we analyzed the expression of ZNF692 in colorectal cancer tissues and adjacent normal
tissues (Figure 1). The results showed that among 248 colorectal cancer specimens, 126 had high expression (IHC score greater
than or equal to 2), 57 had low expression (IHC score less than
2), and 66 had no expression (score 0). The expression level of
ZNF692 in tumors is much higher than that in adjacent normal tissues (Figure 1E), 1.492 ± 0.07091 VS 0.2727 ± 0.1408.
The relationship between ZNF692 expression and pathological features in colorectal cancer patients
The relationship between ZNF692 expression and clinicopathological characteristics of colorectal cancer patients was further
analyzed. The results showed that there were no significant differences between patients with low ZNF692 expression and those
with high ZNF692 expression in terms of gender, T stage, N stage,
neural and/or vascular invasion. (P>0.05, Table 1). However, there
were significant differences in age, M stage and degree of differentiation between the two groups (P<0.05, Table 1). The proportion of high expression of ZNF692 for younger patients was significantly greater than older patients (62.7% vs. 42.0%, P=0.001). M1
stage patients with a higher rate for high expression of ZNF692
compared to M0 stage patients (68.4% vs. 48.1%, P=0.021). Middle/high grade patients showed a higher rate of ZNF692 high expression (55.8% vs. 38.6%, P=0.021).
The effect of ZNF692 expression on patient survival
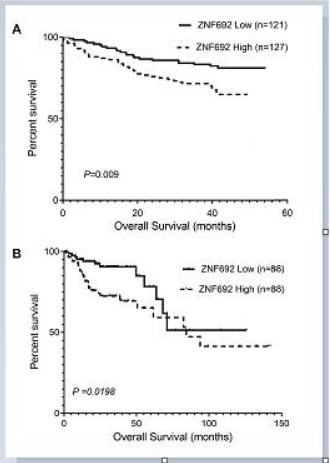
Kaplan-Meier survival curve display that relative higher expression of ZNF692 was associated with inferior overall survival. The
average survival months of patients with low ZNF692 expression
was 47.58±1.36 months, while the survival months of patients
with high ZNF692 expression was 38.83 ± 1.57 months. The difference between the two groups was statistically significant (Figure
2A, P=0.009). Furthermore, The Cox risk regression model was
used to analyze whether ZNF692 protein expression can be used
as an independent predictor of prognosis in colorectal cancer patients. Multivariate analysis showed that ZNF692 expression, age,
gender, and M status were independent predictors of prognosis in
colorectal cancer patients. As shown in Table 2.
Validation of the effect of ZNF692 on overall survival of patients with colorectal cancer in TCGA public data
We downloaded the data of 512 patients with colorectal cancer from TCGA database, 176 of whom had the data required for
survival analysis and were used to draw Kaplan-Meier survival
curve. The results showed that the survival rate of patients with
high ZNF692 expression was significantly lower than that of patients with low ZNF692 expression (Figure 2B, P=0.0198).
Table 1: Correlation between ZNF692 expression and clinicopathological characteristics of CRC.
| Characteristics |
Cases |
ZNF692 expression |
P value |
| Low |
High |
| Gender |
|
|
|
0.82 |
| male |
133 |
64 |
69 |
|
| female |
115 |
57 |
58 |
|
| Age(mean=63) |
|
|
|
0.001* |
| <63 |
110 |
41 |
69 |
|
| ≥ 63 |
138 |
80 |
58 |
|
| Nerve & Vessel invasion |
|
|
|
0.49 |
| No |
140 |
71 |
69 |
|
| Yes |
108 |
50 |
58 |
|
| T status |
|
|
|
0.078 |
| T1-2 |
37 |
23 |
14 |
|
| T3-4 |
211 |
98 |
113 |
|
| N status |
|
|
|
0.849 |
| N0 |
144 |
71 |
73 |
|
| N1/2/3 |
104 |
50 |
54 |
|
| M status |
|
|
|
0.021* |
| M0 |
210 |
109 |
101 |
|
| M1 |
38 |
12 |
26 |
|
| Histologic grade# |
|
|
|
0.039* |
| Low |
44 |
27 |
17 |
|
| Middle/high |
197 |
87 |
110 |
|
* P < 0.05; # The Histologic grade of 7 samples is missing
Table 2: Univariate and multivariate analysis for overall survival
(Cox proportional hazards regression model).
| Risk factors |
Univariate |
Multivariate |
| HR |
P value |
95% CI |
HR |
P value |
95% CI |
| ZNF692 expression
(low/high) |
1.988 |
0.01* |
1.18-3.36 |
1.859 |
0.034* |
1.05-3.30 |
| Age (<63/³63) |
2.364 |
0.003* |
1.33-4.19 |
2.884 |
0.001* |
1.57-5.29 |
| Gender (male/female) |
0.68 |
0.149 |
0.40-1.15 |
0.688 |
0.026* |
0.31-0.93 |
| T status (T1-2/T3-4) |
5.939 |
0.013* |
1.45-24.32 |
2.253 |
0.128 |
0.73-12.73 |
| N status (N0/N1-2) |
2.577 |
<0.001* |
1.53-4.34 |
0.414 |
0.326 |
0.75-2.37 |
| Nerve & Blood
invasion(No/Yes) |
1.297 |
0.16 |
1.05-1.60 |
1.631 |
0.171 |
0.85-2.59 |
| Differentiation |
0.722 |
0.3 |
0.39-1.34 |
0.639 |
0.181 |
0.33-1.23 |
| (moderate &well/poor) |
| M status (M0/M1) |
10.027 |
<0.001* |
5.98-16.82 |
7.491 |
<0.001* |
4.27-13.14 |
HR; Hazard ratio, *P<0.05
Discussion
Our study found that ZNF692 may play a regulatory role as an
oncogene in the progression of colorectal cancers. Because we
observed that ZNF692 is highly expressed in more than half of
the colorectal cancer tissues, this is consistent with the reported
results [6]. And the high expression of ZNF692 is closely related to
the appearance of distant metastasis and the low degree of tumor
differentiation. It is well known that distant metastasis and poor
differentiation are important factors for poor tumor prognosis.
Our survival analysis also confirmed that the survival of patients
with high expression of ZNF692 was worse than that of patients
with low expression. In fact, ZNF692 does promote tumor growth,
Xing Y et al. reported that ZNF692 promotes colorectal adenocarcinoma cell growth and metastasis by activating the PI3K/AKT
pathway [6]. However, the study was only validated from in vitro
cytology tests, and they have not been validated on a large scale
in clinical specimens and lack data for survival analysis.
By analyzing The Cancer Genome Atlas (TCGA) dataset, Zhang
Q et al. confirmed ZNF692 as a potential oncogene in cervical cancer and promotes proliferation, migration and invasion of cervical
cancer cells in vitro [12]. In order to verify the reliability of our results, we also performed a validation analysis of colorectal cancer
public data in the TCGA database. In the analysis, we found that
ZNF692 did affect the survival of patients.
At present, there are few experimental studies and bioinformatics analysis data on ZNF692, and the signal transduction
pathway involved in ZNF692 is still not clear enough. Although
our findings are the first to combine bioinformatics with clinical
specimen data, it is only preliminary to clarify that ZNF692 may play a regulatory role in the progression of colorectal cancer, but
the upstream and downstream molecules of the ZNF692 molecule involved are still unclear. Limited by experimental conditions
and funding, we did not conduct cytological experiments to verify
the molecular mechanism of ZNF692 that involved in colorectal
cancer regulation, nor did we conduct large-scale public data validation of colorectal cancer, including cross-platform multi-group
data integration analysis. Therefore, in the future, based on the
existing results, we will continue to analyze the expression of
ZNF692 in various cancer tissues, including serum, and actively
explore the molecular mechanism of ZNF692 regulating the occurrence and development of colorectal cancer. By elucidating the
role of ZNF692 in the occurrence and development of CRC, it may
provide potential molecules for the target therapy or screening
of patients with CRC, which has very important theoretical and
clinical significance.
Statements & declarations
Funding: This work was supported by Youth Fund of National
Natural Science Foundation of China (Grant No. 81502042) and
Natural Science Foundation for Young Scholars of Jiangsu Province, China (Grant No. BK20140171), and Top Talent Support Program for young and middle-aged people of Wuxi Health Committee (Grant No. HB2020056).
Competing interests: The authors declare that they have no
financial interests.
Author contributions: Xiaosong Ge designed and carried out
experiments, analyzed and interpreted part of the data, Xiaosong
Ge drafted the manuscript. Fen Liu, Xiaoyuan Liu, Xiang Gao and
Yong Mao analyzed and interpreted part of the data. We thanks
Dr. Fang Wang for conducting the TCGA data analysis. All authors
read and approval the final manuscript.
Ethics approval: The study was approved by the Ethics Committee of the Affiliated Hospital of Jiangnan University.
Consent to participate: Informed consent was obtained from
all individual participants included in the study.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, et al.
Global cancer statistics 2020: GLOBOCAN estimates of incidence
and mortality worldwide for 36 cancers in 185 countries. Ca-a Cancer Journal for Clinicians. 2021; 71: 209-249.
- Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the
global cancer statistics 2020. Chin Med J (Engl). 2021; 134: 783-791.
- Heuchan GN, Lally PJ, Beeken RJ, Fisher A, Conway RE. Perception
of a need to change weight in individuals living with and beyond
breast, prostate and colorectal cancer: a cross-sectional survey. J
Cancer Surviv. 2023.
- Mei WJ, Mi M, Qian J, Xiao N, Yuan Y, et al. Clinicopathological
characteristics of high microsatellite instability/mismatch repair-deficient colorectal cancer: A narrative review. Front Immunol.
2022; 13: 1019582.
- Inoue E, Yamauchi J. AMP-activated protein kinase regulates
PEPCK gene expression by direct phosphorylation of a novel zinc finger transcription factor. Biochem Biophys Res Commun. 2006;
351: 793-799.
- Xing Y, Ren S, Ai L, Sun W, Zhao Z, et al. ZNF692 promotes colon adenocarcinoma cell growth and metastasis by activating the PI3K/
AKT pathway. Int J Oncol. 2019; 54: 1691-1703.
- Shirai T, Tanioka Y, Furusho T, Yamauchi J. A Nuclear Factor Involved
in Transcriptional Regulation of the AREBP Gene. J Nutr Sci Vitaminol (Tokyo). 2017; 63: 430-432.
- Zhang Q, Zheng X, Sun Q, Shi R, Wang J, et al. ZNF692 promotes
proliferation and cell mobility in lung adenocarcinoma. Biochem
Biophys Res Commun. 2017; 490: 1189-1196.
- Guo Y, Sheng QH, Li J, Ye F, Samuels DC, Shyr Y. Large Scale Comparison of Gene Expression Levels by Microarrays and RNAseq Using TCGA Data. Plos One. 2013; 8.
- Liu JF, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, et al. An
Integrated TCGA Pan-Cancer Clinical Data Resource to Drive High-Quality Survival Outcome Analytics. Cell. 2018; 173: 400-416.
- Wu J, Wang F, Liu X, Zhang T, Liu F, et al. Correlation of IDH1 and
B7H3 expression with prognosis of CRC patients. Eur J Surg Oncol.
2018; 44: 1254-1260.
- Zhu B, Pan Y, Zheng X, Zhang Q, Wu Y, et al. A clinical, biologic and
mechanistic analysis of the role of ZNF692 in cervical cancer. Gynecol Oncol. 2019; 152: 396-407.